Program
Lectures are from 9 AM to noon, Practicals from 2 to 5 PM.
- Monday : Introduction and overview of IDPs: from theory to practice
- Monday-bis : ≤ 5 min student introductions
- Tuesday : Sampling the conformational space, structural, thermodynamic and kinetic modelling
- Wednesday : Connecting conformational ensembles to experimental results
- Thursday : Phase separation in the cell
- Friday : Novel algorithms for exploring/characterizing high-dimensional conformational spaces
- ...departure: Friday session ends early to allow bus to Ajaccio airport
Links to lecture notes and practicals info will be added to each day as needed.
Monday, 23 Nov: Peter Tompa
Peter Tompa, Vrije Universiteit Brussels, Belgium
Introduction and overview of IDPs: from theory to practice
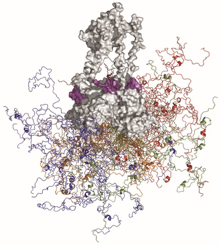
A general introduction to intrinsically disordered proteins through a historical perspective to the most recent developments.
In the theory part, we will cover:
- brief history and basic concepts
- biophysical methods for the characterization of structural disorder
- functions and functional classification of disordered proteins
- structural disorder in diseases: can we target IDPs?
In the practical part, we will look into data and bioinformatics approaches
to structural disorder, such as:
- databases of disordered proteins: DisProt, MobiDB, PED and PhaSePro
- bioinformatics tools: prediction of structural disorder and comparison of disordered ensembles
- low-complexity sequences, prion-like domains and bioinformatics of phase-separating proteins
Monday, 23 Nov, after the practical: Participants Session
Short presentations (≤5 min) to tell about your research interests
Please have a couple of slides (pdf format) ready to project.
Tuesday, 24 Nov: Gerhard Hummer
Gerhard Hummer, Max Planck Institute for Biophysics, Frankfurt am Main, Germany
Sampling the conformational space, structural, thermodynamic and kinetic modelling
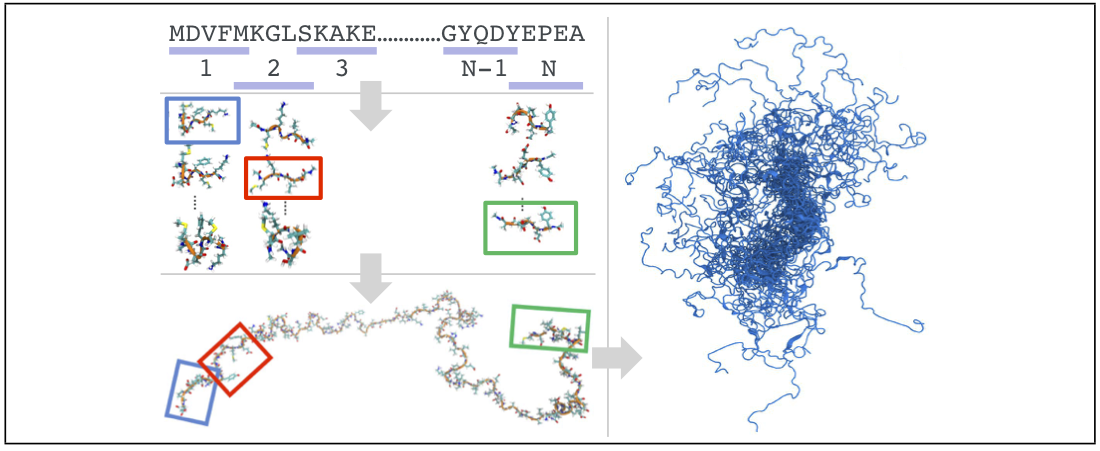
[Pietrek, Stelzl, & Hummer, J. Chem. Theory Comput. 2020]
Description of morning lecture and afternoon practical session coming soon!
Wednesday, 25 Nov: Martin Blackledge and Nathalie Sibille
Martin Blackledge, Institut de Biologie Structurale, Grenoble, France
Nathalie Sibille, Centre de Biochimie Structurale de Montpellier, France
Connecting conformational ensembles to experimental results
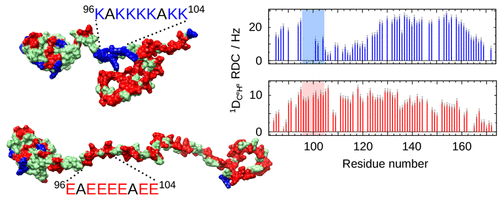
Description of morning lecture and afternoon practical session coming soon!
Thursday, 26 Nov: Jeetain Mittal
Jeetain Mittal, Dept Chemical and Biomolecular Engineering, Lehigh Univ, Pennsylvania
Phase Separation in the Cell

The morning lecture and the afternoon practical will cover physics-based computational models that we have been developing in close collaboration with experimental collaborators for the last ten years.
The lecture will provide a broad overview of available atomistic and coarse-grained models and their relative benefits for specific research problems. A particular emphasis will be a discussion of models suitable for studying the liquid-liquid phase separation of proteins and nucleic acids.
The practical will cover the software and scripts necessary to use our coarse-grained modeling framework capable of providing molecular-level details on the sequence-determinants of protein phase separation due to specific disease mutations and the inclusion of a folded domain, and variation of chain length.
Friday, 27 Nov: Juan Cortés and Frédéric Cazals
Juan Cortés, CNRS LAAS, Toulouse, France
Frédéric Cazals, Inria, Sophia-Antipolis
Novel algorithms for exploring and characterizing high-dimensional conformational spaces

Part I:
- Modeling proteins in internal coordinates: a robotics-inspired approach
- Tripeptides as basic elements to model highly-flexible proteins and regions
- Structural propensity prediction from IDP sequences based on a structural database of tripeptides
- Generation of conformational ensemble models of IDPs/IDRs
Part II:
- Distance based structural measures: least RMSD and combined RMSD
- Nearest neighbor graphs of conformations and energy landscape models
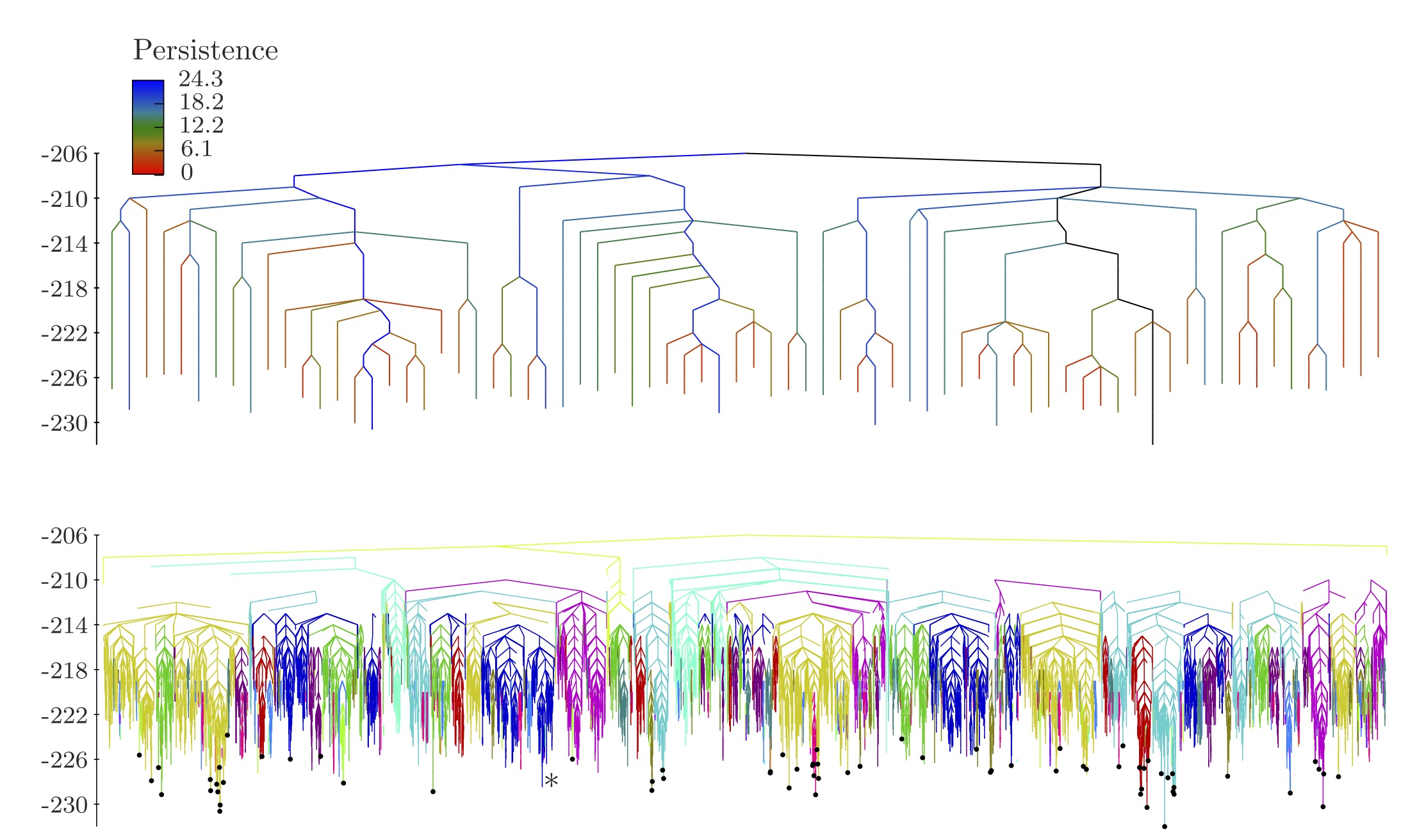
- Persistence based analysis of energy landscapes
- Applications to IDPs
References:
- Predicting secondary structure propensities in IDPs using simple
statistics from three-residue fragments; A. Estaña, A. Barozet,
A. Mouhand, M. Vaisset, C. Zanon, P. Fauret, N. Sibille, P. Bernadó,
J. Cortés; Submitted.
- Realistic ensemble models of intrinsically disordered proteins using a
structure-encoding coil database; A. Estaña, N. Sibille, E. Delaforge,
M. Vaisset, J. Cortés, P. Bernadó; Structure, 27(5): 381-391.e2, 2019
- A reinforcement-learning-based approach to enhance exhaustive protein
loop sampling; A. Barozet, K. Molloy, M. Vaisset, T. Siméon,
J. Cortés; Bioinformatics, 36(4): 1099-1106, 2020
- Characterizing molecular flexibility by combining lRMSD measures;
F. Cazals, and R. Tetley; Proteins, 87 (5), 2019.
- Energy landscapes and persistent minima; J. Carr, and D. Mazauric, and
F. Cazals, and D. Wales; The Journal of Chemical Physics, 144 (5),
2016
- Conformational Ensembles and Sampled Energy Landscapes: Analysis and
Comparison; F. Cazals, and T. Dreyfus, and D. Mazauric, and A. Roth,
and C.H. Robert; J. Comp. Chem., 36 (16), 2015.
Friday afternoon: Bus back to the Ajaccio airport following the practical session.